Advancement of Structure Learning Methods for Time Series Data
Authors
Valen Potloff
Tyler Gibbs
Biwei Huang
Babak Salimi
Introduction
This project extends recent advancements in causal discovery to time series data with instantaneous and time lagged effects. These algorithms can be used to discover not just the size of an effect but also the direction of causality from time series data. We adapt 2 algorithms, GOLEM and DAGMA, to time series and compare it to a similar existing algorithm DYNOTEARS. We have named our adaptations GOLEMTS(GOLEM Time Series) and DAGMATS(DAGMA Time Series).
Causal Discovery
Causal discovery is the term for learning to direction of a causal effect from data. While we might know variables A and B are correlated determining which causes the other or if they are the result of a third effect is much more difficult. The methods discussed here will only consider the case where the casual graph forms a Directed Acyclic Graph(DAG), where no causal loops form. A variety of tools have been developed to solve this problem, recently algorithms that formulate this as a continuous optimisation problem have been an active area of research. NOTEARS first formulated this as a continuous constrained optimization task using a hard DAG constraint.
Unfortunately much of the data we would like to find causal relationships in is very large and the computationally intensive nature of known algorithms limits the impact of this field in active research. However in many situations researchers are dealing with time series data where they are looking for both instant and time lagged effects. If we can incorporate assumptions about this data into these algorithms that dramatically narrow the range of data we need to perform the most computationally intensive calculations we can see significant improvements to our analysis.
DYNOTEARS
In 2020 NOTEARS was adapted to the time series data with the creation of DYNOTEARS. This divided the data into two sections, instantaneous effects and time lagged effects. Time lagged effects can not be affected by variables measured in the future, incorporating these assumptions into the algorithm allowed to to more accurately and much more quickly process datasets. We take this approach and extend it to GOLEM and DAGMA. Since GOLEM and DAGMA are significantly more accurate than DYNOTEARS on large datasets our implementations of these algorithms for time series datasets has seen a similar improvement over DYNOTEARS.
We provide an implementation of DYNOTEARS primarily to serve as a baseline to compare GOLEMTS and DAGMATS as they are superior in nearly every circumstance.
GOLEMTS
GOLEM built on NOTEARS’ success by showing that with a better formulation of the likelihood function and a soft sparsity constraint the hard dag constraint could be weakened to a soft constraint. This unconstrained optimization problem was much easier to solve. We extend the likelihood function developed for GOLEM for time lagged effects resulting in GOLEMTS. The specifics of how this works mathematically can be found in the report.pdf in the linked github repository.
GOLEM has two separate implementations, GOLEM_EV assumes equal variances among the variables while GOLEM_NV does not, we mirror this in our implementation with GOLEMTS_EV and GOLEMTS_NV.
DAGMATS
DAGMA incorporates assumptions about how matrix representations of DAGs form M-matrixes to create a DAG constraint with better performing gradients which allow standard optimisation methods to reach the true DAG representing the causal effects faster and more accurately. We apply this new constraint to the objective function we developed for the previous methods to get DAGMATS. Again the specifics of how this is accomplished can be found in report.pdf.
Results
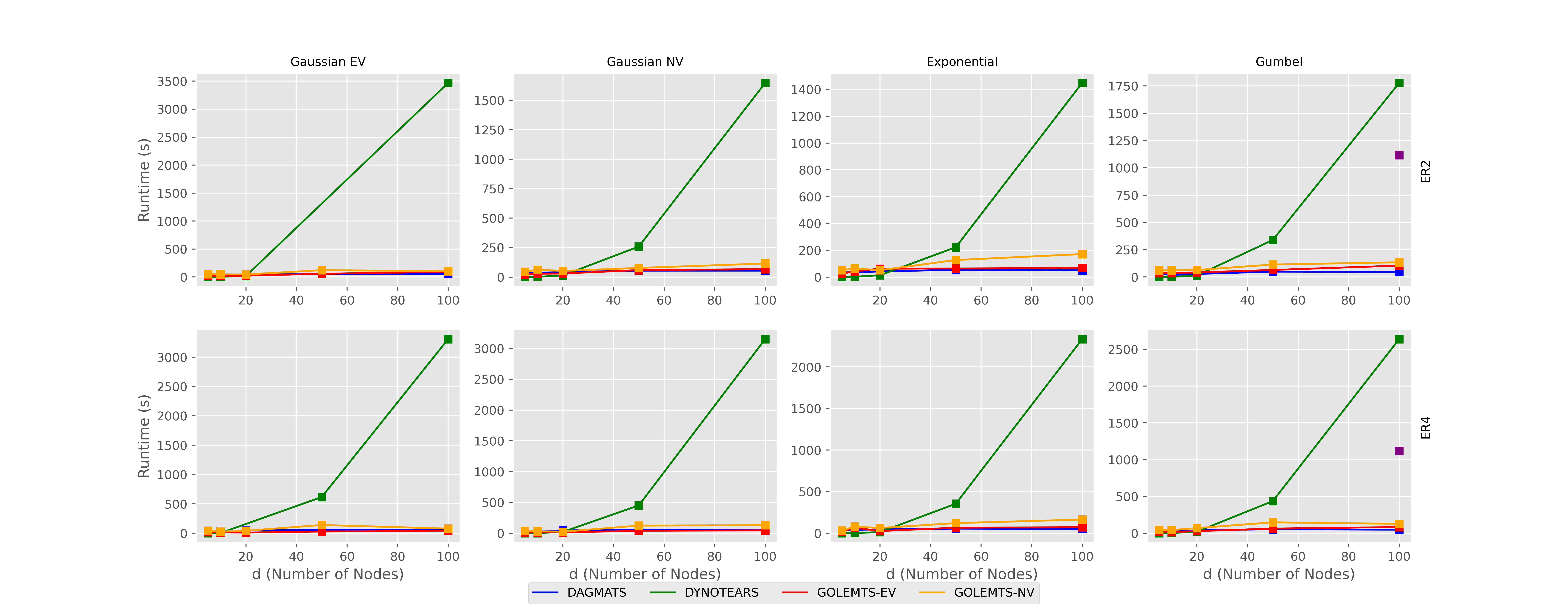
The following graphs show the relative performance of DYNOTEARS, GOLEM_EV, GOLEM_NV, and DAGMATS with datasets of increasing size generated with equal variance gaussian noise, nonequal variance gaussian noise, exponential noise, and gumball noise.
First we examine the runtime and see that GOLEM_EV, GOLEM_NV, and DAGMATS perform significantly better on large datasets than DYNOTEARS.
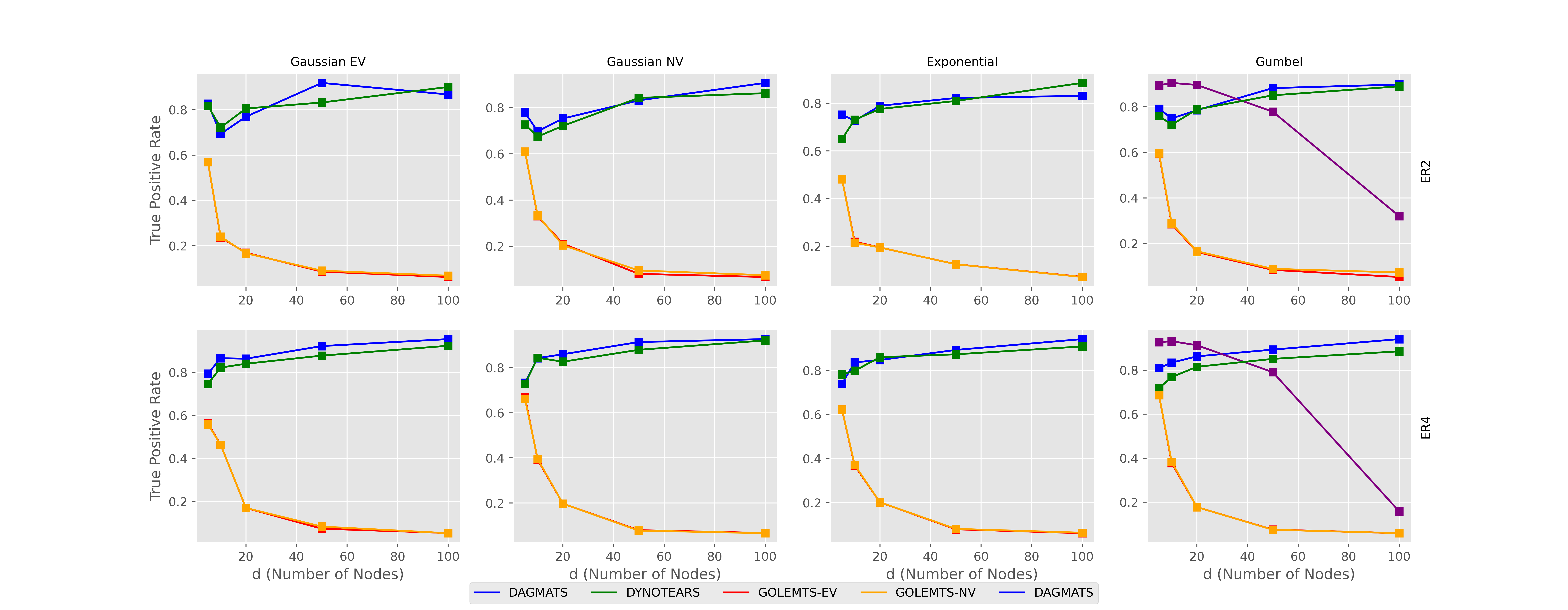
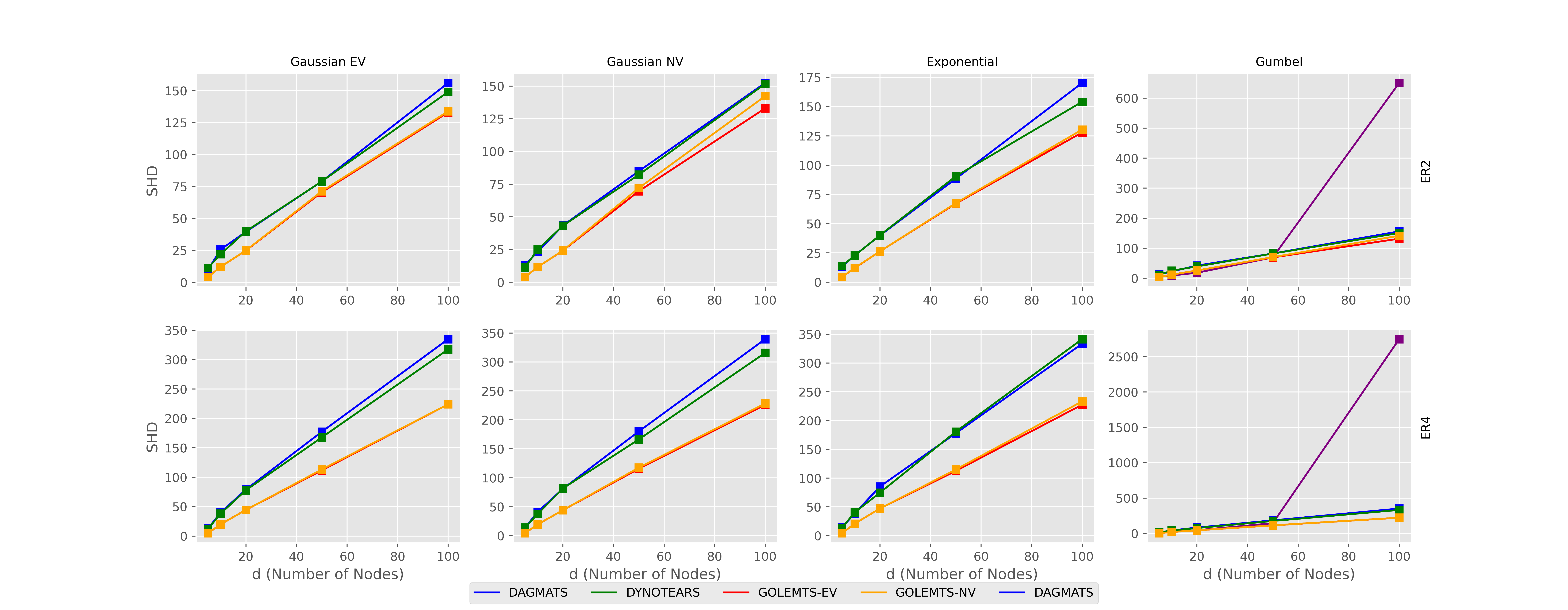
There are many ways to measure error when finding causal graphs. We will look at two, the true positive rate and structural hamming distance.
The true positive rate is exactly what it sounds like. The percent of predicted links that are true, a higher number is better.
Structural hamming distance is the number of additions or removals required to change our predicted graph to the true graph. Lower is better.
First we examine the true positive rate and see a significant divergence where we see that on larger datasets DYNOTEARSTS and DAGMATS maintain their precision but GOLEMTS suffers significantly.
However this story is complicated when we also look at structural hamming distance where both implementations of GOLEMTS outperform both DAGAMTS and and DYNOTEARS, although by a more modest degree.
Usage
Run your own
We have provided implementations of DYNOTEARS GOLEMTS and DAGMATS. You can run these on your own datasets or on Synthetic datasets generated by our implementation. We provide two primary methods from running these algorithms, from the main method and from a call to the underlying python function.
main.py
You can run each method on generated data or a provided csv file using ‘python main.py’ command from the root directory
‘python main.py’ is followed by the name of the method (DYNOTEARS, GOLEMTS, DAGMATS) followed by arguments specifying the data and output directory.
-p is the number of different times slots in the data and must always be specified
If you are using existing data you give the filename of the csv file under –data as shown below
python3 main.py GOLEMTS \
--data filename.csv \
-p 1
If no –out parameter is specified it will write to output.json. If the –out parameter is specified it will write to that file. The output is a json object containing the original casual graph if known and the estimated causal graph, allowing the user to analyze the accuracy.
python3 main.py GOLEMTS \
--data filename.csv \
-p 1 \
--out new_output.json
It can also generate synthetic data using the following two parameters
- -n number of observations
- -d number of variables
- –degree number of causal relationships to model
The method will then be ran on the synthetic data and the estimated graph will be written to the output file
python3 main.py GOLEMTS \
-n 10 \
-d 3 \
-p 1 \
--degree 4
python3 main.py DAGMATS \
-n 10 \
-d 3 \
-p 1 \
--degree 4 \
python3 main.py DYNOTEARS \
-n 10 \
-d 3 \
-p 1 \
--degree 4 \
--out new_output.json
Direct Python Calls
This method provides the highest degree of customisation. Each function in formated to take the number of observations(n), number of variables per timeslot(d), number of timeslots(p) followed by the data(Y) formatted with n rows and d*p columns. After these positional arguments are the particular methods hyperparameters as discussed in the methods section in the introduction. Last is the number of iterations to run(epochs) before returning the best estimate.
-
src.train.run_DYNOTEARS(n, d, p, Y, w_thresh=0.01, epochs=100):
-
src.train.run_GOLEMTS(n, d, p, Y, lambda_1=0.1, lambda_2=1, ev=True, lr=3e-3, lambda_3=9, epochs=1000, warmup_epochs=0):
-
src.train.run_DAGMATS(n, d, p, Y, lambda1=0.01, lambda2=0.03, lr=0.02, w_threshold=0, epochs=1000)
Reference
Ng, Ignavier, AmirEmad Ghassami, and Kun Zhang. “On the role of sparsity and dag constraints for learning linear dags.” Advances in Neural Information Processing Systems 33 (2020): 17943-17954.
Zheng, Xun, et al. “Dags with no tears: Continuous optimization for structure learning.” Advances in neural information processing systems 31 (2018).
Bello, Kevin, Bryon Aragam, and Pradeep Ravikumar. “Dagma: Learning dags via m-matrices and a log-determinant acyclicity characterization.” Advances in Neural Information Processing Systems 35 (2022): 8226-8239.
Pamfil, Roxana, et al. “Dynotears: Structure learning from time-series data.” International Conference on Artificial Intelligence and Statistics. PMLR, 2020.